-Search query
-Search result
Showing all 35 items for (author: kinoshita & c)

EMDB-36048: 
Cryo-EM structure of hZnT7-Fab complex in zinc-unbound state, determined in outward-facing conformation
Method: single particle / : Han BB, Inaba K, Watanabe S

EMDB-36049: 
Cryo-EM structure of hZnT7-Fab complex in zinc-bound state, determined in outward-facing conformation
Method: single particle / : Han BB, Inaba K, Watanabe S

EMDB-36050: 
Cryo-EM structure of hZnT7-Fab complex in zinc-unbound state, determined in heterogeneous conformations- one subunit in an inward-facing and the other in an outward-facing conformation
Method: single particle / : Han BB, Inaba K, Watanabe S

EMDB-36051: 
Cryo-EM structure of hZnT7-Fab complex in zinc state 2, determined in heterogeneous conformations- one subunit in an inward-facing zinc-bound and the other in an outward-facing zinc-bound conformation
Method: single particle / : Han BB, Inaba K, Watanabe S

EMDB-36052: 
Cryo-EM structure of hZnT7DeltaHis-loop-Fab complex in zinc-unbound state, determined in outward-facing conformation
Method: single particle / : Han BB, Inaba K, Watanabe S

EMDB-36053: 
Cryo-EM structure of hZnT7DeltaHis-loop-Fab complex in zinc-bound state, determined in outward-facing conformation
Method: single particle / : Han BB, Inaba K, Watanabe S

EMDB-36055: 
Cryo-EM structure of hZnT7-Fab complex in zinc state 1, determined in heterogeneous conformations- one subunit in an inward-facing zinc-bound and the other in an outward-facing zinc-unbound conformation
Method: single particle / : Han BB, Inaba K, Watanabe S

PDB-8j7t: 
Cryo-EM structure of hZnT7-Fab complex in zinc-unbound state, determined in outward-facing conformation
Method: single particle / : Han BB, Inaba K, Watanabe S

PDB-8j7u: 
Cryo-EM structure of hZnT7-Fab complex in zinc-bound state, determined in outward-facing conformation
Method: single particle / : Han BB, Inaba K, Watanabe S

PDB-8j7v: 
Cryo-EM structure of hZnT7-Fab complex in zinc-unbound state, determined in heterogeneous conformations- one subunit in an inward-facing and the other in an outward-facing conformation
Method: single particle / : Han BB, Inaba K, Watanabe S

PDB-8j7w: 
Cryo-EM structure of hZnT7-Fab complex in zinc state 2, determined in heterogeneous conformations- one subunit in an inward-facing zinc-bound and the other in an outward-facing zinc-bound conformation
Method: single particle / : Han BB, Inaba K, Watanabe S

PDB-8j7x: 
Cryo-EM structure of hZnT7DeltaHis-loop-Fab complex in zinc-unbound state, determined in outward-facing conformation
Method: single particle / : Han BB, Inaba K, Watanabe S

PDB-8j7y: 
Cryo-EM structure of hZnT7DeltaHis-loop-Fab complex in zinc-bound state, determined in outward-facing conformation
Method: single particle / : Han BB, Inaba K, Watanabe S

PDB-8j80: 
Cryo-EM structure of hZnT7-Fab complex in zinc state 1, determined in heterogeneous conformations- one subunit in an inward-facing zinc-bound and the other in an outward-facing zinc-unbound conformation
Method: single particle / : Han BB, Inaba K, Watanabe S

EMDB-32407: 
RNA polymerase II elongation complex bound with Elf1 and Spt4/5, stalled at SHL(-4) of the nucleosome
Method: single particle / : Osumi K, Kujirai T, Ehara H, Sekine S, Takizawa Y, Kurumizaka H

EMDB-32408: 
RNA polymerase II elongation complex bound with Elf1 and Spt4/5, stalled at SHL(-3.5) of the nucleosome
Method: single particle / : Osumi K, Kujirai T, Ehara H, Sekine S, Takizawa Y, Kurumizaka H

EMDB-32409: 
RNA polymerase II elongation complex bound with Elf1 and Spt4/5, stalled at SHL(-3) of the nucleosome
Method: single particle / : Osumi K, Kujirai T, Ehara H, Sekine S, Takizawa Y, Kurumizaka H

EMDB-34685: 
RNA polymerase II elongation complex bound with Rad26 and Elf1, stalled at SHL(-3.5) of the nucleosome
Method: single particle / : Osumi K, Kujirai T, Ehara H, Kinoshita C, Saotome M, Kagawa W, Sekine S, Takizawa Y, Kurumizaka H

PDB-7wbv: 
RNA polymerase II elongation complex bound with Elf1 and Spt4/5, stalled at SHL(-4) of the nucleosome
Method: single particle / : Osumi K, Kujirai T, Ehara H, Sekine S, Takizawa Y, Kurumizaka H

PDB-7wbw: 
RNA polymerase II elongation complex bound with Elf1 and Spt4/5, stalled at SHL(-3.5) of the nucleosome
Method: single particle / : Osumi K, Kujirai T, Ehara H, Sekine S, Takizawa Y, Kurumizaka H

PDB-7wbx: 
RNA polymerase II elongation complex bound with Elf1 and Spt4/5, stalled at SHL(-3) of the nucleosome
Method: single particle / : Osumi K, Kujirai T, Ehara H, Sekine S, Takizawa Y, Kurumizaka H

PDB-8he5: 
RNA polymerase II elongation complex bound with Rad26 and Elf1, stalled at SHL(-3.5) of the nucleosome
Method: single particle / : Osumi K, Kujirai T, Ehara H, Kinoshita C, Saotome M, Kagawa W, Sekine S, Takizawa Y, Kurumizaka H

EMDB-34430: 
Cryo-EM structure of the human RAD52 protein
Method: single particle / : Kinoshita C, Takizawa Y, Saotome M, Ogino S, Kurumizaka H, Kagawa W

PDB-8h1p: 
Cryo-EM structure of the human RAD52 protein
Method: single particle / : Kinoshita C, Takizawa Y, Saotome M, Ogino S, Kurumizaka H, Kagawa W

EMDB-30360: 
34-fold symmetry salmonella MS ring
Method: single particle / : Kawamoto A, Miyata T, Makino F, Kinoshita M, Minamino T, Imada K, Kato T, Namba K

EMDB-30361: 
11 fold-syymetry of salmonella MS ring
Method: single particle / : Kawamoto A, Miyata T, Makino F, Kinoshita M, Minamino T, Imada K, Kato T, Namba K

EMDB-30363: 
Without imposing symmetry of salmonella MS ring
Method: single particle / : Kawamoto A, Miyata T, Makino F, Kinoshita M, Minamino T, Imada K, Kato T, Namba K

EMDB-30613: 
Structure of the bacterial flagellar basal body
Method: single particle / : Kawamoto A, Miyata T, Makino F, Kinoshita M, Minamino T, Imada K, Kato T, Namba K

EMDB-30940: 
Salmonella flagella MS-ring protein FliF 1-503 having C33 symmetry
Method: single particle / : Makino F, Kawamoto A, Namba K

EMDB-30941: 
Salmonella flagella MS-ring protein FliF 1-456 having C35 symmetry
Method: single particle / : Makino F, Kawamoto A, Namba K

EMDB-30942: 
Salmonella flagella MS-ring protein FliF 1-456 having C34 symmetry
Method: single particle / : Makino F, Kawamoto A, Namba K
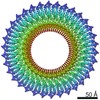
EMDB-30612: 
34-fold symmetry Salmonella S ring formed by full-length FliF
Method: single particle / : Kawamoto A, Miyata T

PDB-7d84: 
34-fold symmetry Salmonella S ring formed by full-length FliF
Method: single particle / : Kawamoto A, Miyata T, Makino F, Kinoshita M, Minamino T, Imada K, Kato T, Namba K

EMDB-30378: 
Salmonella FliF-FliG ring complex
Method: single particle / : Kawamoto A, Kato T, Kinoshita M, Minamino T, Namba K

EMDB-30379: 
11-fold symmetry of Salmonella FliF-FliG ring complex
Method: single particle / : Kawamoto A, Kato T, Kinoshita M, Minamino T, Namba K
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
